Avoid solid
CAD Modeling
6
Posts
2
Posters
1.8k
Views
2
Watching
-
Hi,
I am modeling some neural pathways and portions of it go along the muscle layer in the human body model. Is it possible to avoid solid intersection?
I came across something like this in the IMSafe toolbox:
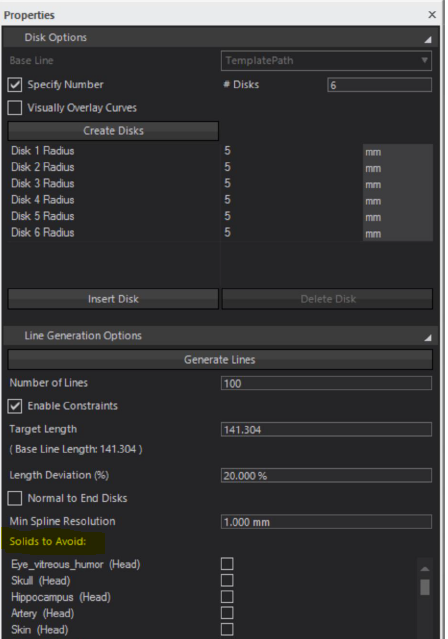
So I'm wondering if it is possible to do this using Python.
Thanks!
-
Here is a script I found. Not sure it still runs, but I guess it should help you get started:
import tempfile import os import s4l_v1.model as model from s4l_v1.model import Vec3, Unit import s4l_v1.simulation as simulators import s4l_v1.simulation.emfdtd as emfdtd # Preamble --------------- import s4l_v1.document as doc sim = doc.AllSimulations[0] # assumptions sphere = model.AllEntities()["Sphere 1"] assert sphere assert isinstance(sim, emfdtd.Simulation) sim_xtor = sim.Results() # pin this variable, otherwise wrapped members are deleted efield_port = sim_xtor["Overall Field"]["EM E(x,y,z,f0)"] hfield_port = sim_xtor["Overall Field"]["EM H(x,y,z,f0)"] # Demo --------------------------------------------------- from s4l_v1.analysis.em_evaluators import ImplantSafety from s4l_v1.analysis import RegisterAlgorithm # Creates ImSafe algorithm imsafe = ImplantSafety() # 0. create algorithm and connect to e-field port imsafe.SetInputConnection(0, hfield_port) # Disks options base_line = model.CreateSpline([Vec3(52.0, 0.0, 0.0), Vec3(0, 32, 0), Vec3(-9, 70, 0)]) imsafe.SetBaseLine(base_line) # 1. define baseline total_len = imsafe.EvalBaseLineLength() imsafe.ModelUnits = Unit.MilliMeter imsafe.SetDisks( position=[0, 0.1, 1], radii=[10.0] * 3 ) # 2. define disks along baseline centers, normals, radii = imsafe.GetDisks() print("centers", centers, imsafe.ModelUnits) print("normals", normals, imsafe.ModelUnits) print("radii", radii, imsafe.ModelUnits) # Line generation options #3. setup line options imsafe.NumberOfLinesProp.Value = 5 imsafe.LengthDeviationProp.Value = 10 # % imsafe.NormalToEndDisksProp.Value = True imsafe.MinSplineResolutionProp.Value = 0.01 * total_len # or # imsafe.MinSplineResolutionProp.SetValueAs(0.05, Unit.Meter) imsafe.EnableConstraintsProp.Value = False # 4. setup obstacles imsafe.SetAsObstacles([sphere]) RegisterAlgorithm(imsafe) assert imsafe.UpdateAttributes(), "Guarantees output ports are updated" imsafe.ClearOutputLines() assert imsafe.Update(0) filepath = os.path.join(tempfile.mkdtemp(), "line.txt") print("Exporting to", filepath) imsafe.ExportLines(filepath, snapshot_index=0) imsafe.ClearOutputLines() imsafe.ImportLines(filepath) # Creates viewer import s4l_v1.analysis.viewers as viewers dv = viewers.SurfaceViewer() dv.SetInputConnection(0, imsafe.GetOutputPort(2)) RegisterAlgorithm(dv) -
- it simply takes the first simulation. Note, the evaluator needs an existing simulation because it tests if the splines pass through voxels of the "Entities to Avoid" list
- the entities to avoid are set via
SetAsObstacles(entity_list). the script assumes this entity is called "Sphere `" - the script creates a spline as baseline, but you could also create the baseline in the GUI, and get it via
base_line = model.AllEntities()["My Baseline"]