Introducing the Image Modeling module: Image and LabelField
-
If you are working with medical image data, or some other form of 3D images, and image segmentations, the Sim4Life Python provides a set of tools which allow batch processing, and powerful custom operations based on Python scripts and thirdparty modules such as numpy, vtk, etc.
The image model entity is defined in the ImageModeling module. The api includes the properties and methods:
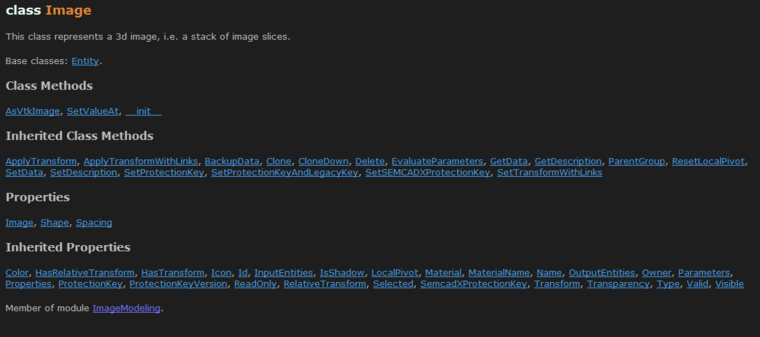
The interesting properties are Image, which returns a 3D numpy array that wraps the actual image data of the entity, and the Shape, Spacing and Transform. If you are missing the typical 'Origin' property, this information is stored in the Transform.
ImageModeling.LabelField is a class which stores 3D segmentation data, i.e. it is an (16-bit unsigned integer) image with information about the labels (label index, name and color).
The following simple script illustrates how to load an iSEG segmentation (LabelField) and generate a surface model using the Python API.
import XCoreModeling import ImageModeling import ViP ents = XCoreModeling.Import(r"F:\Data\test_iseg.prj") label_fields = [e for e in ents if isinstance(e,ImageModeling.LabelField)] assert len(label_fields)==1, "expecting one LabelField" print "There are", len(label_fields[0].Labels), "labels" print "Label of Artery is", label_fields[0].Labels["Artery"] print "The segmentation has dimensions", label_fields[0].Shape print "The segmentation has spacing", label_fields[0].Spacing print "The segmentation has orientation/offset", label_fields[0].Transform ViP.ExtractSurface(label_fields[0], smooth=True, simplify=True)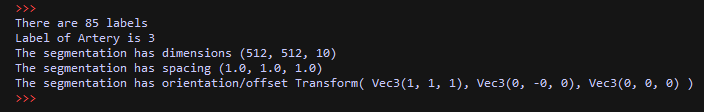
Note about s4l_v1 API: Above classes and methods are partly already contained in the official Sim4Life Python API (s4l_v1), or will be added in the coming release within the s4l_v1.model.image scope.